# install.packages('ggsignif')
library(ggplot2)
library(ggsignif)
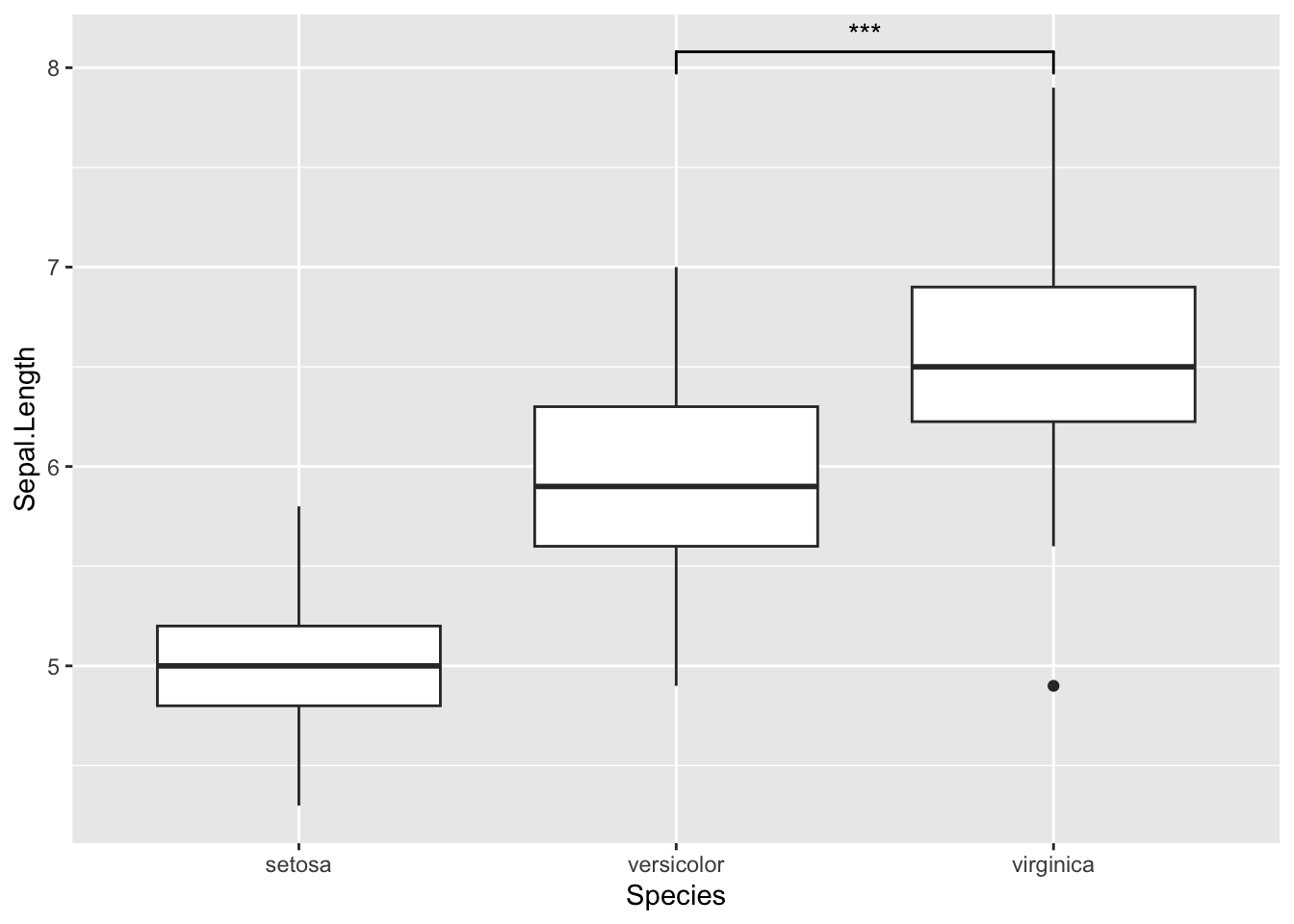
library(data.table)ggplot(iris, aes(x = Species, y = Sepal.Length)) +
geom_boxplot() + # using `ggsignif` to display comparison of interest
geom_signif(
comparisons = list(c("versicolor", "virginica")),
map_signif_level = TRUE
)
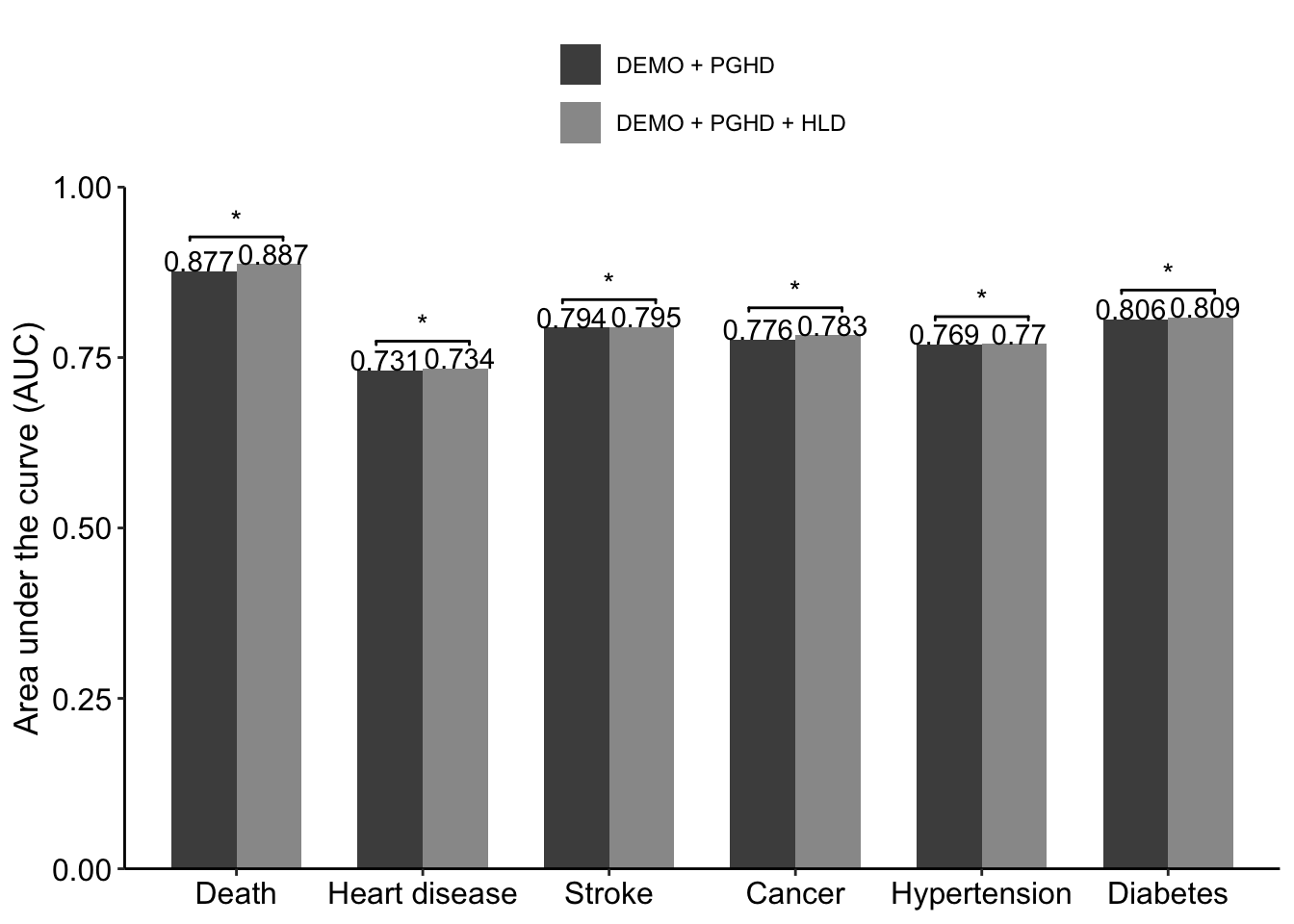
dt_male <- data.table(
outcome = rep(c('Death','Heart disease','Stroke',
'Cancer','Hypertension','Diabetes'),2),
model = as.factor(rep(2:3, each=6)),
value = c(0.877,0.731,0.794,0.776,0.769,0.806,
0.887,0.734,0.795,0.783,0.770,0.809)
)
dt_male$outcome <- factor(dt_male$outcome,
levels=c('Death','Heart disease','Stroke','Cancer','Hypertension','Diabetes'))
setkey(dt_male, outcome)
sig_loc <- c()
for(i in seq(1,11,2)){
sig_loc <- c(sig_loc,mean(dt_male$value[i:i+1]))
}
dt_male |>
ggplot(aes(x=outcome, y=value, fill=model))+
geom_col(position = position_dodge(),
width = .7) +
theme_classic() +
scale_y_continuous(limits=c(0,1),
expand=c(0,0)) +
scale_fill_manual(values=c('grey30','grey60'),
labels=c('DEMO + PGHD',
'DEMO + PGHD + HLD'),
) +
theme(legend.position = 'top',
axis.title.x = element_blank(),
axis.title.y = element_text(size=13),
axis.text = element_text(size=12, color='black'),
# axis.ticks.x = element_line(color=c(rep(NA,len-1), rep('black',len))),
legend.title = element_blank(),
legend.direction = 'vertical') +
guides(fill=guide_legend(byrow = T))+ # legend 간격 띄우기
labs(y='Area under the curve (AUC)') +
geom_text(aes(label=value, y=value+0.015),
position = position_dodge(width=0.8)) +
# 유의미한 변수 표시
geom_signif(
y_position = sig_loc+0.04,
xmin= c(0.75, 1.75, 2.75, 3.75, 4.75, 5.75),
xmax = c(1.25,2.25, 3.25, 4.25, 5.25, 6.25),
annotations = c('*')
)
레퍼런스
- https://const-ae.github.io/ggsignif/articles/intro.html